Abstract:
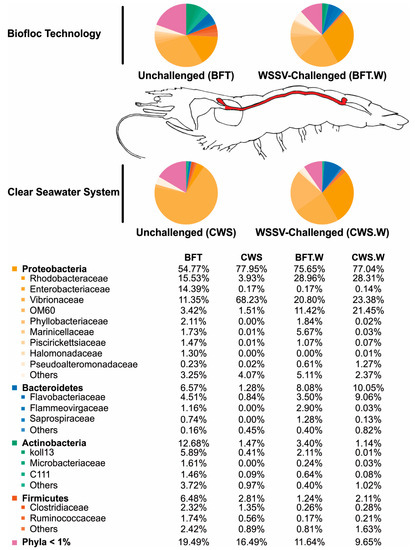
We provide a global overview of the intestinal bacteriome of Litopenaeus vannamei in two rearing systems and after an oral challenge by the White spot syndrome virus (WSSV). By using a high-throughput 16S rRNA gene sequencing technology, we identified and compared the composition and abundance of bacterial communities from the midgut of shrimp reared in the super-intensive biofloc technology (BFT) and clear seawater system (CWS). The predominant bacterial group belonged to the phylum Proteobacteria, followed by the phyla Bacteroidetes, Actinobacteria, and Firmicutes. Within Proteobacteria, the family Vibrionaceae, which includes opportunistic shrimp pathogens, was more abundant in CWS than in BFT-reared shrimp. Whereas the families Rhodobacteraceae and Enterobacteriaceae accounted for almost 20% of the bacterial communities of shrimp cultured in BFT, they corresponded to less than 3% in CWS-reared animals. Interestingly, the WSSV challenge dramatically changed the bacterial communities in terms of composition and abundance in comparison to its related unchallenged group. Proteobacteria remained the dominant phylum. Vibrionaceae was the most affected in BFT-reared shrimp (from 11.35 to 20.80%). By contrast, in CWS-reared animals the abundance of this family decreased from 68.23 to 23.38%. Our results provide new evidence on the influence of both abiotic and biotic factors on the gut bacteriome of aquatic species of commercial interest.
Figure: